Research
|
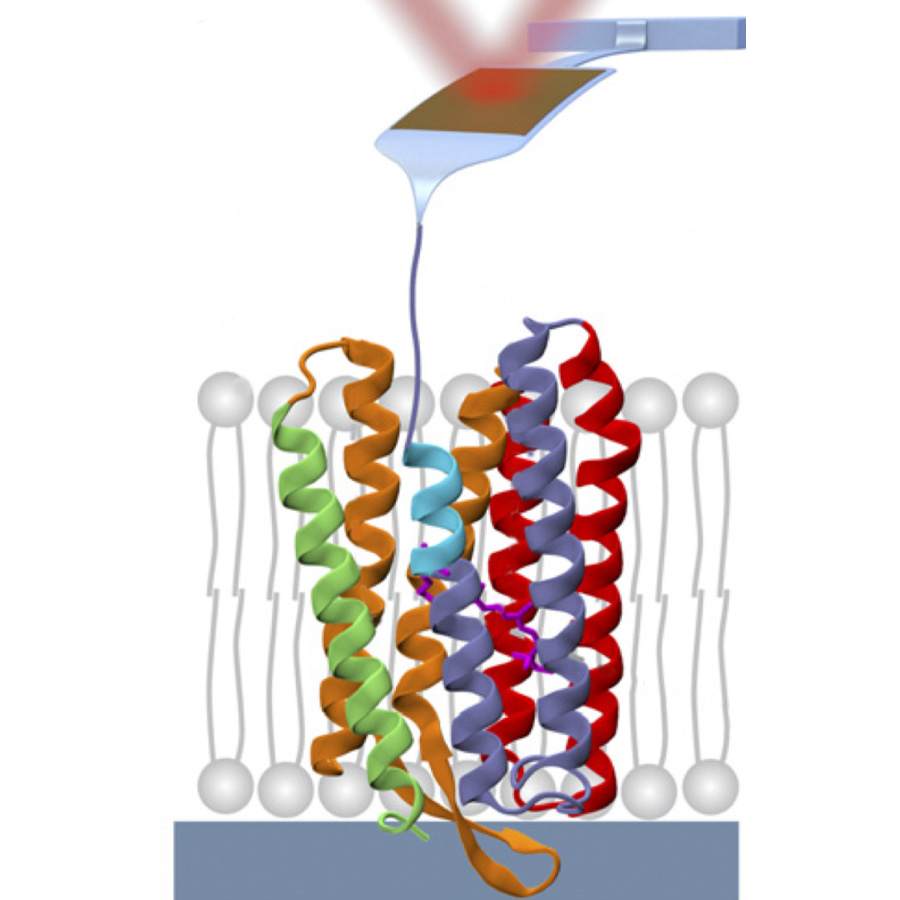
Single-molecule studies of membrane proteinProteins embedded in the cell membrane are key to many biological processes and are the targets of many drugs because they contact both the cytoplasm and the extracellular environment. Understanding how membrane proteins adopt correctly folded structures, and how those structures are perturbed by ligand binding and disease-causing mutations, requires the ability to measure the energetics of the interactions holding them together. Traditional biochemical approaches, however, are limited in biological interpretability and in what proteins can be studied. Building on my postdoctoral work with Tom Perkins at JILA, our lab is interested in using precise forces applied by atomic force microscopy (AFM) to reversibly unfold and refold small segments of membrane proteins (e.g., cyan in figure) to make thermodynamically well-defined measurements of their energetics as a function of biologically relevant parameters such as ligand concentration, introduction of mutations, and lipid environment. |